Welcome to the website for The Plotkin Research Group
We do research at the interface of biology and physics.
Our current projects involve the molecular-genetic study of the evolution of multicellularity, protein misfolding in neurodegenerative disease, and more recently, viral evolution and molecular therapeutics to SARS-CoV-2. Please see our Research Page for more details.
Plotkin Lab News:
Congratulations to Santanu Sasidharan on receiving the Michael Smith Health Research BC Trainee Award!
Recently Awarded Grants:
- PROGENITER: Pathogen Response Optimization by GENeratIng ThErapeutics Rationally, Canada Biomedical Research Fund Award: PROGENITER will bolster Canada’s pandemic preparedness by creating a suite of ready-to-deploy antibody treatments for current health threats like COVID-19 and influenza, while positioning Canada to mount a rapid response to novel threats as they emerge. Canada Biomedical Research Fund Award: $14.46 million, Biosciences Research Infrastructure Fund Award: $16.40 million
- CIHR: Priority Announcement: Research on Prevention or Response to Covid-19 or Similar Future Pandemics: “A platform for developing mutation-resistant vaccines and antibodies for future viral variants, using SARS-CoV-2 as a model”
- Genome BC sequencing funding for SARS-CoV-2 research at Plotkin Lab
- CIHR: Priority announcement funding: Pandemic Preparedness and Health Emergencies Research: “A development platform for mutation-resistant vaccines and antibodies for SARS-CoV-2 future variants”
Recent Publications:
- Kaplan J, Gibbs E, Coutts JA, Zhao B, Mackenzie IR, Plotkin SS, Cashman NR “Relationship between efficacy and preferential targeting of soluble Aβ aggregates” Alzheimer’s Dement. 11:e70184 (2025)
- Antibodies targeting Rack1and Methods of Use Thereof, US Provisional Patent 63/840,423, Inventors: Neil R Cashman, Steven S Plotkin, Ching-Chung Hsueh, Beibei Zhao, Filing date: July 8, 2025
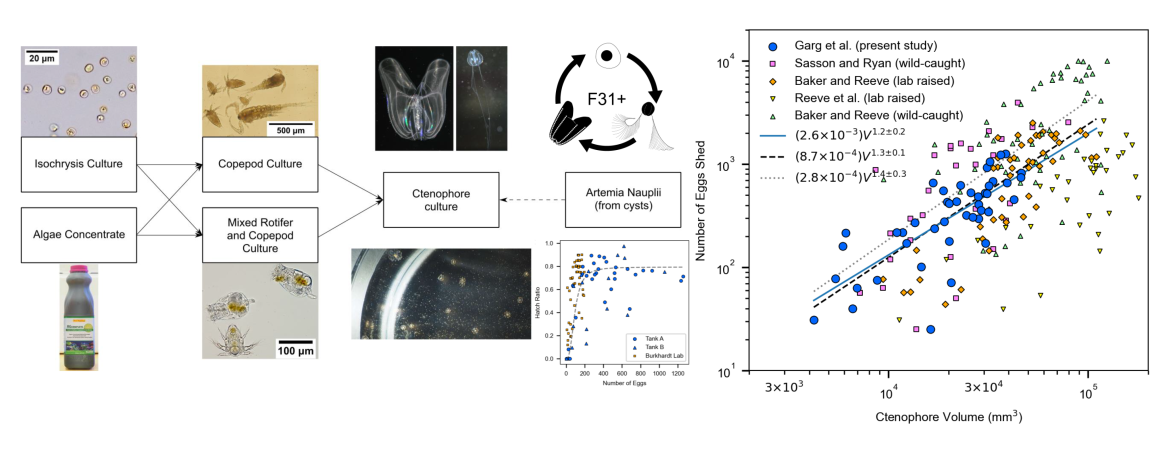
- Garg P , Frey C, Browne WB, and Plotkin SS, “Reproductive success of inbred strain MV31 of the ctenophore Mnemiopsis leidyi in a self-sustaining inland laboratory culture system” Heliyon Volume 11, Issue 4, e42542 (2025)
- McAlary L, Nan JR, Shyu C, Sher M, Plotkin SS, Cashman NR, “Amyloidogenic regions in beta-strands II and III modulate the aggregation and toxicity of SOD1 in living cells” Open Biol.14: 230418 (2024)
- Pokrishevsky E, DuVal MG, McAlary L, Louadi S, Pozzi S, Roman A, Plotkin SS, Dijkstra A, Julien JP, Allison WT, Cashman NR “Tryptophan Residues in TDP-43 and SOD1 Mediate the Cross-seeding and Toxicity of SOD1” Journal of Biological Chemistry, Vol 300, Issue 5:107207 (2024)
- Garg P, Hsueh SCC, Plotkin SS, “Testing the feasibility of targeting a conserved region on the S2 domain of SARS-CoV-2 spike protein” Biophysical Journal, Vol 123, Issue 8, P992-1005 (2024) Cover article
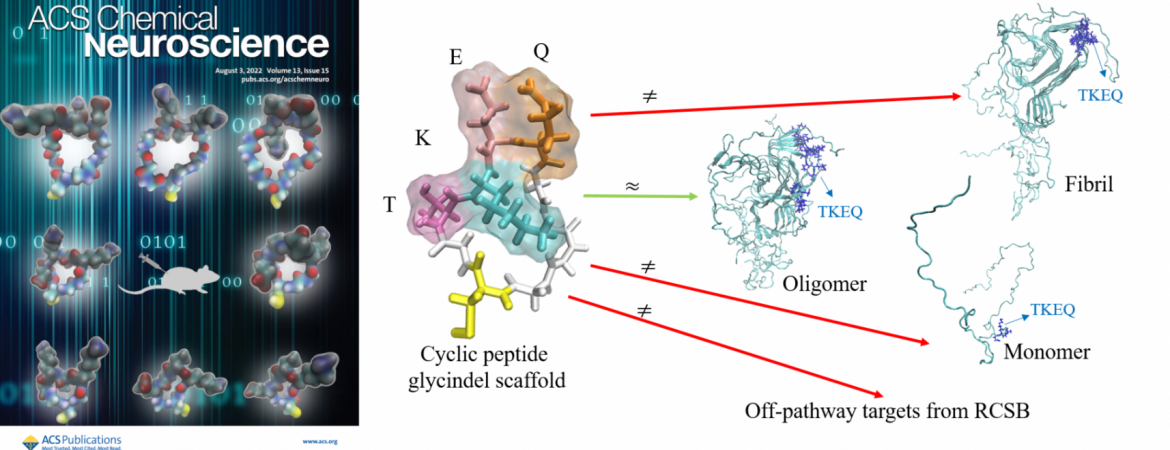
- Hsueh SCC, Nijland M, Aina A; Plotkin SS, “Cyclization Scaffolding for Improved Vaccine Immunogen Stability: Application to Tau Protein in Alzheimer’s Disease” Journal of Chemical Information and Modeling 64, 6, 2035–2044 (2024)
- Cyclic compound/peptide comprising an A-beta15-18 peptide and a linker that is covalently coupled to the n-terminus residue and the c-terminus residue of the A-BETA15-18 peptide, US Patent 11,970,522, Inventors: Neil R Cashman, Steven S Plotkin, Filing date: April 30, 2024
- Conserved Epitopes in Coronaviruses and Vaccines and Antibodies Thereto. US Provisional Patent Application No.: US 63/447,146, Inventors: Steven S Plotkin, Pranav Garg, Ching-Chung Hsueh, Santanu Sasidharan. Full Application Filing date: February 21, 2024
Recent Submissions:
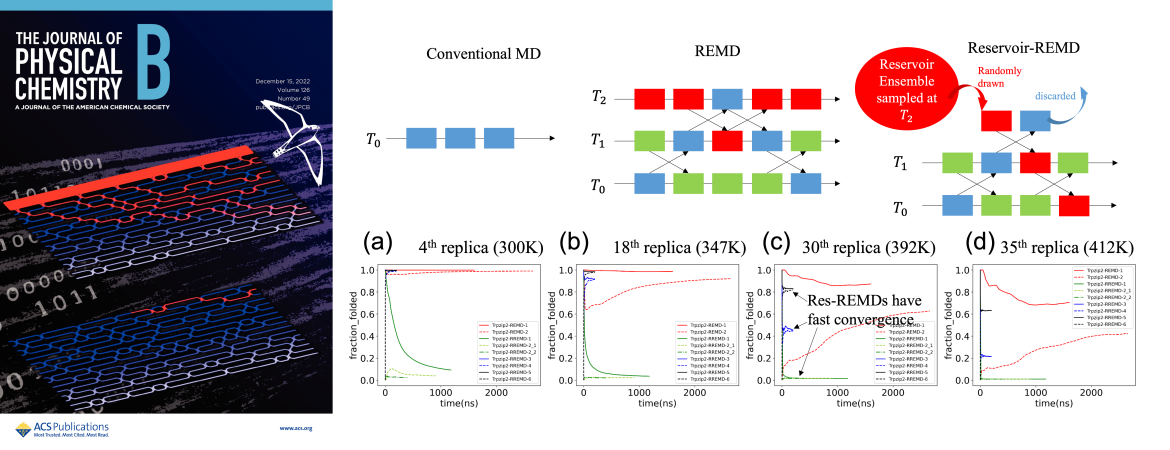
- Hsueh SCC and Plotkin SS, “Reservoir-REMD facilitates kinetic rescue from metastable peptide conformations” (in review)
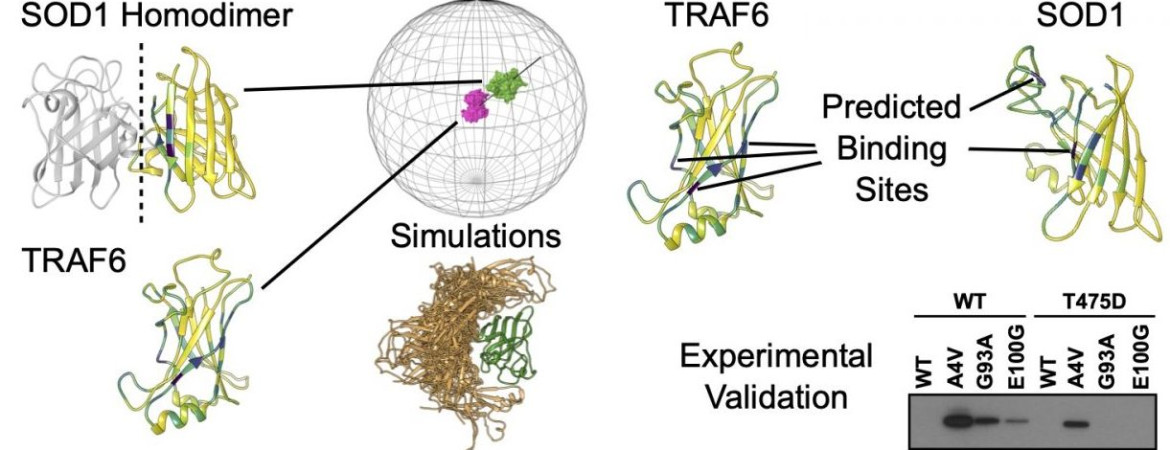
- DuVal MG, McAlary L, Habibi M, Garg P, Sher M, Cashman NR, Allison WT and Plotkin SS “Physical Properties of Novel Aggressive and Beneficial SOD1 Mutants Predict Phenotypes Consistent with ALS” (submitted, bioRxiv)
Recent Recorded Presentations:
We are grateful to the Digital Technology Supercluster, Canadian Institutes of Health Research, Genome BC, Mitacs, and Westgrid/Compute Canada for supporting our research.